Effective Deep Learning Workflow on HPC
Overview
Teaching: 15 min
Exercises: 50 minQuestions
How do we train and tune deep learning models effectively using HPC?
How to convert a Jupyter notebook to a Python script?
How can we perform post-analysis of HPC computations using Jupyter?
Objectives
Switch the mindset from a single-tasked model development workflow to the ‘dispatch and analyze’ mode which offloads heavy-duty computations to HPC.
Perform a full conversion of a Juptyer Notebook to a Python script.
Analyze and aggregate results from HPC model tuning jobs on Jupyter.
Introduction
Motivation
In the previous episode, we introduced the model tuning procedure on a Jupyter notebook. As you may recall, the process was painfully time-consuming, because we have to wait for one training to complete before we can start another model training. Not only this leads to long wait time to finish all the required trainings, this approach will also be impractical in real-world deep learning, where each model training process could take hours or even days to complete. In this episode, we will introduce an alternative workflow to tune a deep learning model on an HPC system. With HPC, model trainings can be submitted and executed in parallel. We will show that this approach can greatly reduce the total human time needed to try out all the various hyperparameter combinations.
There are several potential issues with utilizing Juptyer Notebook, especially with well established neural
network code.
One is that code must by executed one cell at a time.
The potential for non-linear execution of blocks/cells of code
is more helpful when developing and testing code, rather
than when running established code.
Another is that the code must be ran from the beginning everytime you close and reopen the notebook
(since the variable values are not saved between runs).
Another is that Jupyter Notebook cannot run multiple runs (e.g., network trainings)
in parallel.
Running multiple runs is difficult in Jupyter Notebook, since each run needs to be queued
one after another, even though there is no dependency relationship between runs.
For these reasons, sometimes it is more efficient to utilize Python and batch scripting
instead of relying on Jupyter Notebook.
Specifically, switching to Python script will improve throughput and turnaround time
for the Baseline_Model notebook introduced in a prior episode.
Before switching, make sure to establish a working pipeline for machine learning.
Batch Scheduler
One huge benefit from converting a Jupyter notebook into a Python script, is because it allows for the use of batch (non-interactive) scheduling (of the Python scripts). It allows the user to launch neural network trainings through one batch (SLURM) script. Real machine-learning work requires many repetitive experiments, each of which may take a long time to complete. The batch scheduler allows many experiments to be carried out in parallel, allowing for more and faster results.
HPC is well suited for this type of workflow – in fact, it is the most effective when used in this way. Here are the key components of the “batch” way of working:
-
A job scheduler (such as SLURM job scheduler on HPC) to manage the jobs and run them on the appropriate resources;
-
The machine learning script written in Python, which will read inputs from files and write outputs to files and/or standard output;
-
The job script to launch the machine learning script in the non-interactive environment (e.g. HPC compute node);
-
A way to systematically repeat the experiments with some variations. This can be done by adding some command-line arguments for the (hyper)parameters that will be varied for each experiment.
Since the jobs (where each job is one or more experiments) will be run in parallel, keep in mind the following:
-
The Python script will need to utilize the same input file(s), but each one must work in their own work directory. This will assist in organization and post-processing analysis. This will also assist in ensuring that there is no clashing amongst parallel jobs/experiments.
-
Using proper job names. Each experiment should be assigned a unique job name. This job name is very useful for organization, troubleshooting, and post-processing analysis.
The SLURM script can be modified to combine the two ideas. It can pass the unique job name to the Python script. The Python script can then create a working directory that includes the (unique) job name.
A Baseline Model to Tune on HPC
In this episode, we will demonstrate the process of converting a Jupyter notebook
to a Python script using the baseline neural network model for the sherlock_18apps
classification.
We have a Jupyter notebook prepared, Baseline_Model.ipynb,
which contains a complete pipeline of machine learning from data loading and
preparation, followed by neural network model definition, training, and saving.
The codes in this notebook are essentially the same as those that define the
baseline model in the previous episode of this lesson
(“[Tuning Neural Network Models for Better Accuracy][NN-ep25-model-tuning]”).
The saved model can be reloaded later to deploy it for the actual application.
The Baseline Model
As a reminder, the baseline model for tuning the
sherlock_18appsclassifier is defined with the following hyperparameters:
- one hidden layer with 18 neurons
- learning rate of 0.0003
- batch size of 32
Steps to Convert a Jupyter Notebook to a Python Script
The first step is to convert the Jupyter notebook to a Python script. There are several ways to convert a Juypter Notebook into a Python script:
-
Manual process: Go over cell-by-cell in the Jupyter interface, copying and pasting the relevant code cells to a blank Python script. (Both Jupyter Lab and Jupyter Notebook support editing a Python script.) This can be especially useful when there is a lot of convoluted code or if there are multiple iterations of the same code in the same notebook. While this allows for very intentional and precise selection of code segments, it can be time consuming and prone to manual errors.
-
Automatic conversion: Use the
jupyter nbconvertcommand. This command extracts all the codes in a given notebook into a Python script. The script will generally need be edited to account for the differences between the interactive Jupyter platform and noninteractive execution in Python.
When using wahab, make sure that nbconvert
is loaded in (you can module load tensorflow-cpu/2.6.0) crun.
This is an example that converts the Baseline_Model.ipynb to Basline_Model.py using nbconvert.
crun jupyter nbconvert --to script Baseline_Model.ipynb
Cleaning up the Code
If selecting to use the nbconvert option, make sure to make adjustments to clean up the
code and make corrections.
-
Remove comments such as
In[1]:,In[2], etc., which is used to note the separation of the cells. -
Verify the retaining of all comments. It also comments out the text blocks created in the Jupyter notebook.
-
Remove any unnecessary (code) cells that have been commented out.
-
Remove any Jupyter notebook exclusive things, such as
%matplotlib inline. -
Verify the retention of any commands used in Jupyter notebook to view information. Some commands in Jupyter allow the user to view information, such as
head()andtail(), but these will not print from the Python script without being surrounded byprint().
Also, note that the previously saved cell outputs are not included (not even as a comment). This is fine, since it is the output from a previous run.
Editing and Adjusting the Code
Remove all interactive and GUI input/output. Input prompts should be modified to read the input file.
Any outputs should be saved to a unique working directory and/or be uniquely named.
This mindfulness will assist in allowing the output to be machine processable later.
This includes images - changing matplotlib.show() to savefig()
and other valuable outputs (e.g. tables) - saved as files (e.g. CSV).
Example Using nbconvert: Leading to Baseline_Model.py
Exercise: Converting Baseline_Model.ipynb to Baseline_Model.py
1. Utilize the nbconvert command explained above.
Solution
crun jupyter nbconvert --to script Baseline_Model.ipynb2. Remove all unnecessary comments in comment header and import statement sections. Also, remove the unnecessary Jupyter notebook lines.
Solution
#!/usr/bin/env python # coding: utf-8 # # 1. Loading Python Libraries import os import sys import pandas as pd import numpy as np import matplotlib.pyplot as plt import matplotlib.gridspec as gridspec # CUSTOMIZATIONS (optional) np.set_printoptions(linewidth=1000) # tools for machine learning: import sklearn from sklearn import preprocessing from sklearn.model_selection import train_test_split # for evaluating model performance from sklearn.metrics import accuracy_score, confusion_matrix # classic machine learning models from sklearn.linear_model import LogisticRegression from sklearn.tree import DecisionTreeClassifier # tools for deep learning: import tensorflow as tf import tensorflow.keras as keras # Import key Keras objects from tensorflow.keras.models import Sequential from tensorflow.keras.layers import Dense from tensorflow.keras.optimizers import Adam #RUNIT # For developers only: import tensorflow.keras.models as _keras_models import tensorflow.keras.layers as _keras_layers3. Clean up the load sherlock sections.
Remove the unnecessary comments to clean up the code. The cell separation comments and the RUNIT comments.
Solution
# # 2. Loading Sherlock Applications Data df = pd.read_csv("../sherlock/sherlock_18apps.csv", index_col=0) ## Summarize the dataset print("* shape:", df.shape) print() print("* info::\n") df.info() print() print("* describe::\n") print(df.describe().T) print() df.head(10) df.tail(10)4. Clean up the cleaning of the data section.
Remove the unnecessary cell separation comments.
Solution
"""Perform cleaning of a Sherlock 19F17C dataset. All the obviously bad and missing data are removed. """ # Missing data or bad data del_features_bad = [ 'cminflt', # all-missing feature 'guest_time', # all-flat feature ] df2 = df.drop(del_features_bad, axis=1) print("Cleaning:") print("- dropped %d columns: %s" % (len(del_features_bad), del_features_bad)) print("- remaining missing data (per feature):") isna_counts = df2.isna().sum() print(isna_counts[isna_counts > 0]) print("- dropping the rest of missing data") df2.dropna(inplace=True) print("- remaining shape: %s" % (df2.shape,)) """Separate labels from the features""" print("Step: Separating the labels (ApplicationName) from the features.") labels = df2['ApplicationName'] df_features = df2.drop('ApplicationName', axis=1)5. One-hot encoding and feature scaling
Delete the unnecessary comments, such as the cell separation comments and optional comment.
Solution
"""Perform one-hot encoding for **all** categorical features.""" print("Step: Converting all non-numerical features to one-hot encoding.") df_features = pd.get_dummies(df_features) df_features.head() df_features.info() """Step: Feature scaling using StandardScaler.""" print("Step: Feature scaling with StandardScaler") df_features_unscaled = df_features scaler = preprocessing.StandardScaler() scaler.fit(df_features_unscaled) # Recast the features still in a dataframe form df_features = pd.DataFrame(scaler.transform(df_features_unscaled), columns=df_features_unscaled.columns, index=df_features_unscaled.index) print("After scaling:") print(df_features.head(10)) print()6. Train-val split section
Remove the comments that separate the cells and the extraneous RUNIT comments.
Solution
"""Step: Perform train-val split on the master dataset. This should be the last step before constructing & training the model. """ val_size = 0.2 random_state = 34 print("Step: Train-val split val_size=%s random_state=%s" \ % (val_size, random_state)) train_features, val_features, train_labels, val_labels = \ train_test_split(df_features, labels, test_size=val_size, random_state=random_state) print("- training dataset: %d records" % (len(train_features),)) print("- valing dataset: %d records" % (len(val_features),)) sys.stdout.flush() print("Now the feature matrix is ready for machine learning!") train_features.head(10) # Both df and df2 work for the devel version, only df2 for learners version # Leaving the scripts as is and using df2 should be fine app_counts = df2.groupby('ApplicationName')['CPU_USAGE'].count() print(app_counts) print("Num of applications:",len(app_counts)) train_L_onehot = pd.get_dummies(train_labels) val_L_onehot = pd.get_dummies(val_labels) train_L_onehot.head()7. Defining the Baseline Model (NN_Model_1H method).
Remove the unnecessary cell separation comments.
Solution
# # 3. The Baseline Model def NN_Model_1H(hidden_neurons, learning_rate): """Definition of deep learning model with one dense hidden layer""" model = Sequential([ # More hidden layers can be added here Dense(hidden_neurons, activation='relu', input_shape=(19,), kernel_initializer='random_normal'), # Hidden Layer Dense(18, activation='softmax', kernel_initializer='random_normal') # Output Layer ]) adam_opt = Adam(lr=learning_rate, beta_1=0.9, beta_2=0.999, amsgrad=False) model.compile(optimizer=adam_opt, loss='categorical_crossentropy', metrics=['accuracy']) return model8. Reproducibility hack, calling the model, and fitting the model.
Remove the unnecessary cell separation comments.
Solution
# Reproducibility hacks! np.random.seed(38477518) tf.random.set_seed(967288341) model_1H = NN_Model_1H(18,0.0003) model_1H_history = model_1H.fit(train_features, train_L_onehot, epochs=10, batch_size=32, validation_data=(val_features, val_L_onehot), verbose=2)9. Plot_loss, plot_acc, and combine_plots methods.
Nothing to change, just make sure the comments noting the separation in cells is deleted.
10. Last section of code that creates (and combines) the plots as well as saves the history, plot, and model.
Nothing to change, just make sure the comments noting the separation in cells is deleted.
SLURM Batch Script Review
To create the SLURM batch script, we need to define the SBATCH directives, module loading, any environmental variables, and then the executable SLURM commands.
The SBATCH directives
This is the section where every line starts with #SBATCH.
These are the first lines of the script (not including the line #!/bin/bash.
We will set the job name to NN_Model_1H and output file name is compiled using
two different SLURM filename patterns.
The job name %x and %j for the job allocation number of the running job.
The partition -p is set to main (the default partition, see Partition Policies).
The job is given a maximum of 1 hour.
Much like python, aside from the SBATCH directives section, anything else with # is treated as a comment.
Module Loading and Environmental Variables
This is the section for module (i.e. package) loading.
In this case, we load the default container_env and tensorflow-gpu/2.6.0 modules.
These work like any other variable in Linux, but must be called using a $.
Both CRUN and CRUN_ENVS_FLAGS variables are used to make the executable line easier to read.
Save this as Baseline_Model.slurm.
#!/bin/bash
#SBATCH -J Baseline_Model
#SBATCH -o %x.out%j
#SBATCH -p main
#SBATCH -t 1:00:00
module load container_env
module load tensorflow-gpu/2.6.0
CRUN=crun.tensorflow-gpu
CRUN_ENVS_FLAGS="-p $HOME/envs/default-tensorflow-gpu-2.6.0"
$CRUN $CRUN_ENVS_FLAGS python3 Baseline_Model.py
Post Analysis on Baseline_Model.py
See post_analysis_Baseline_Model.ipynb, which is recreated below.
Step 0: import modules
import os
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
Step 1: Discovery of all the results
## We know all of the output file names (because we set them)
## So for now, just use the set file paths
csvFile = './model_1H18N_history.csv'
imgPath = "./loss_acc_plot.png"
modelPath = "./model_1H18N.h5"
Step 2: Load the results
# Load in the csv file containing the loss, accuracy, val_loss, and val_accuracy
df = pd.read_csv(csvFile)
print(df)
loss accuracy val_loss val_accuracy
0 1.103675 0.675141 0.548827 0.870166
1 0.407072 0.904734 0.320540 0.924509
2 0.274303 0.931864 0.242521 0.938406
3 0.217764 0.946759 0.199030 0.950527
4 0.181833 0.959201 0.169203 0.962776
5 0.156120 0.966429 0.146968 0.967134
6 0.136310 0.970242 0.129598 0.970760
7 0.120869 0.973940 0.117158 0.973927
8 0.108925 0.976939 0.105800 0.977058
9 0.099547 0.978536 0.097061 0.979219
# Collect the plots for each job.
# Here, we can use matplotlib to import the saveed images.
from matplotlib import pyplot as plt
from matplotlib import image as mpimg
image = mpimg.imread(imgPath)
plt.title("Baseline_Model")
fig = plt.imshow(image)
plt.axis('off')
fig.axes.get_xaxis().set_visible(False)
fig.axes.get_yaxis().set_visible(False)
plt.show()
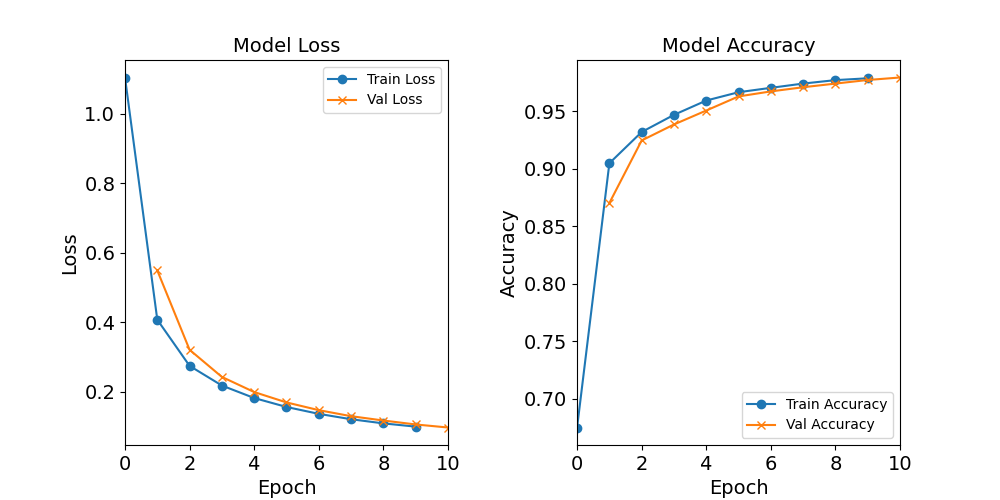
## Load in the model, though for this case, we do not need this
#model_1H = keras.saving.load_model("./model_1H18N.h5")
Validation phase: visualization of training data
1) Inspect if the trainings did not go as expected
2) Visually inspect for anomalies
3) Visually or numerically check for convergence (e.g. check the last 4-5 epochs, what the slope is like; any fluctuations?)
Most of these can be validated using the graph
1) The training did behave as expected. No under or over fitting. Also, the training loss is decreasing as the number of epochs increases, while the accuracy increases.
2) There are no major anomalies (such as any random spikes or dips).
3) It looks like both the accuracy and loss functions are starting to converge.
Therefore, this model behaves as expected.
Analysis phase: Visualizing the results
Since this post-analysis script is for one model’s results, this step can be skipped.
Running Experiments Utilizing Command Line Arguments (NN_Model_1H)
We have just created a Python and SLURM script that runs one model with fixed hyperparameters. However, we want to be able to utilize batch scripts to run multiple experiments at the same time. One way to accomplish this is by utilizing command line arguments. Command line arguments will add the capability of defining hyperparameters in the SLURM file.
Implementing Command Line Arguments
Duplicate Baseline_Model.py and rename the new copy to be NN_Model_1H.py.
Make the following changes to the code.
0) (Optional) Change the heading of the file
"""
NN_Model_1H.py
Python script for model tuning experiments.
Running this script requires four arguments on the command line:
python3 NN_Model_1H.py HIDDEN_NEURONS LEARNING_RATE BATCH_SIZE EPOCHS
"""
1) Define the hyperparameters at the top and assign them
the command line argument values using sys.argv.
We create a standard model output directory name that
is based on the hyperparameter values.
As explained above, having a standard name assists in maintaining the
experiments and during post-analysis.
Next, print the hyperparameters to the output file.
HIDDEN_NEURONS = int(sys.argv[1])
LEARNING_RATE = float(sys.argv[2])
BATCH_SIZE = int(sys.argv[3])
EPOCHS = int(sys.argv[4])
# Create model output directory
MODEL_DIR = "model_1H" + str(HIDDEN_NEURONS) + "N_lr" + str(LEARNING_RATE) + "_bs" + str(BATCH_SIZE) + "_e" + str(EPOCHS)
if not os.path.exists(MODEL_DIR):
os.makedirs(MODEL_DIR)
print()
print("Hyperparameters for the training:")
print(" - hidden_neurons:", HIDDEN_NEURONS)
print(" - learning_rate: ", LEARNING_RATE)
print(" - batch_size: ", BATCH_SIZE)
print(" - epochs: ", EPOCHS)
print()
2) Then, change all (variable) references to the hardcoded hyperparameters.
model_1H = NN_Model_1H(HIDDEN_NEURONS, LEARNING_RATE)
model_1H_history = model_1H.fit(train_features,
train_L_onehot,
epochs=EPOCHS, batch_size=BATCH_SIZE,
validation_data=(val_features, val_L_onehot),
verbose=2)
3) Next, change the output file names using the model directory path created above. This will allow all of the output files to share a common name and be contained in their respective model directory.
history_file = os.path.join(MODEL_DIR, 'model_history.csv')
plot_file = os.path.join(MODEL_DIR, 'loss_acc_plot.png')
model_file = os.path.join(MODEL_DIR, 'model_weights.h5')
metadata_file = os.path.join(MODEL_DIR, 'model_metadata.json')
4) Then, add the additional step of creating the meta data. Using JSON allows the user to easily read and write the structured meta data. It saves name-value pairs that can be queried.
# Because of the terseness of Keras API, we create our own definition
# of a model metadata.
# timestamp of the results (at the time of saving)
model_1H_timestamp = time.strftime('%Y-%m-%dT%H:%M:%S%z')
# last epoch results is a key-value pair (i.e. a Series)
last_epoch_results = history_df.iloc[-1]
model_1H_metadata = {
# Our own information
'dataset': 'sherlock_18apps',
'keras_version': tf.keras.__version__,
'SLURM_JOB_ID': os.environ.get('SLURM_JOB_ID', None),
'timestamp': model_1H_timestamp,
'model_code': '1H18N',
# Hyperparameters
'optimizer': 'Adam',
# the number of hidden layers will be deduced from the length
# of the hidden_neurons array:
'hidden_neurons': [HIDDEN_NEURONS],
'learning_rate': LEARNING_RATE,
'batch_size': BATCH_SIZE,
'epochs': EPOCHS,
# Some results
'last_results': {
'loss': round(last_epoch_results['loss'], 8),
'accuracy': round(last_epoch_results['accuracy'], 8),
'val_loss': round(last_epoch_results['val_loss'], 8),
'val_accuracy': round(last_epoch_results['val_accuracy'], 8),
}
}
with open(metadata_file, 'w') as F:
json.dump(model_1H_metadata, F, indent=2)
These command line arguments can be passed to the Python script via the
SLURM script.
They will be added to the crun line after the .py file argument.
Make sure that the order of the variables in the Python and SLURM script match!
The values (syntax-wise) can be defined a couple different ways.
One way is by doing --[variableName] [value].
Another is by doing ""
Creating SLURM Script with Command Line Arguments
First, duplicate the Baseline_Model.slurm file and rename it to NN_Model_1H.slurm
Then, change the name of the job
#SBATCH -J NN_Model_1H
Then, add the block that will take in the command line arguments. Note, the actual values will be set in a different slurm script, or can be passed within the sbatch command.
# HYPERPARAMS_LIST contains four arguments,
# which must be given in the command-line argument:
# * the number of hidden neurons
# * the learning rate
# * the batch size
# * the number of epochs to train
HIDDEN_NEURONS=$1
LEARNING_RATE=$2
BATCH_SIZE=$3
EPOCHS=$4
Then, add the hyperparameters to the last line.
$CRUN $CRUN_ENVS_FLAGS python3 NN_Model_1H.py "$HIDDEN_NEURONS" "$LEARNING_RATE" "$BATCH_SIZE" "$EPOCHS"
Run NN_Model_1H.slurm by passing in the arguments into the command line.
In the terminal, run the
NN_Model.1H.slurm. In this example, the number of hidden neurons in the first layer is 18, the learning rate is 0.0003, the batch size is 32, and the number of epochs is 10.sbatch NN_Model_1H.slurm 18 0.0003 32 10Since these were the same hyperparameters used in
Baseline_Model.py, the results should look the same.
Using a Master Launcher Script to Launching a Series of Jobs for a Certain Hyperparameter Scanning Task
We can use a master launcher script with a “for loop,” to allow for the launching of a series of jobs for a certain hyperparameter scanning task. This can be used to replicate the experiments from the previous episode.
Create a submit-scan-hidden-neurons.sh file that will submit multiple jobs that vary the number of hidden neurons
(in the first layer).
To do so, define the hyperparameters at the top of the script that do not vary.
Then, create a for loop with the varying hyperparameter with the values to experiment.
Next, define JOBNAME, which will be the name of the output file.
Define the RUNDIR variable to the given convention.
Then, add the additional variables to the sbatch command.
For this script, 8 different jobs will be spawned, each with a
0.0003 learning rate, batch size of 32, and 30 epochs.
Each of the 8 jobs will have a different amount of hidden neurons (in the first layer).
So, the first job will call NN_Model_1H.slurm with a 0.0003 learning rate, batch size of 32, and 30 epochs, and with 1 hidden neuron.
#!/bin/bash
#HIDDEN_NEURONS= -- varied
LEARNING_RATE=0.0003
BATCH_SIZE=32
EPOCHS=30
for HIDDEN_NEURONS in 1 2 4 8 12 18 40 80; do
JOBNAME=model-tuning-1H${HIDDEN_NEURONS}N
RUNDIR=model_1H${HIDDEN_NEURONS}N_lr${LEARNING_RATE}_bs${BATCH_SIZE}_e${EPOCHS}
echo "Training for hyperparams:" "$HIDDEN_NEURONS" "$LEARNING_RATE" "$BATCH_SIZE" "$EPOCHS"
sbatch -J "$JOBNAME" NN_Model_1H.slurm "$HIDDEN_NEURONS" "$LEARNING_RATE" "$BATCH_SIZE" "$EPOCHS"
done
Post-Analysis Results from hidden_neuron_experiments.sh
See post_analysis_NN_Model_1H.ipynb, also reproduced below.
Step 0: Import libraries
import os
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
Step 1: Discovery of all the results
import glob
# Get the directories that contain the results for each model.
# The directory will start with "model_1H".
# If the model ran correctly, it should contain a .csv file in it.
# To do this, use the library glob https://docs.python.org/3/library/glob.html.
# This is sufficient for this step, since each directory will contain its csv, plot, model, and json file.
dirList = []
dirPath = "*/model_1H*N*/"
# check whether the model produced output
dirListTemp = glob.glob(dirPath)
for dirPathI in dirListTemp:
if glob.glob(dirPathI+"*.json"):
dirList.append(dirPathI)
print(dirList)
Step 2: Load the results
1) First, use the json file paths to load in the meta data. This meta data will be used in the data frame to help identify each model. We will accomplish this by using MultiIndex (https://pandas.pydata.org/docs/reference/api/pandas.MultiIndex.html). Using MultiIndex assists with readability of the table, since rows that share the same label only need to have it listed once.
2) Initialize a pre-allocated DataFrame and assign the MultiIndex according to the meta data from each model.
3) Populate the results (training loss, training accuracy, validation loss, and validation accuracy) into the DataFrame.
4) Query the model’s graphic results (loss v. epochs and accuracy v. epochs graphs).
# 2.1: Read in Json files and load meta data.
import json
numRows = []
newIndex = []
for dirI in dirList:
# For each directory, read in the json file and get each of the neccessary MultiIndex values
with open(dirI + "/model_metadata.json") as f:
data = json.load(f)
hidden_neurons = data['hidden_neurons']
learning_rate = data['learning_rate']
batch_size = data['batch_size']
epochs = data['epochs']
jobID = data['SLURM_JOB_ID']
model_type = "model_1H"
numRows.append(epochs)
# Create a tuple with the meta data for model_type, jobID, hidden neurons list (one value for the number of neurons in
# that layer), learning rate, batch size, and then the epochs.
# This will be used to create the MultiIndex object.
newIndex.extend(tuple([(model_type, jobID, str(hidden_neurons), learning_rate, batch_size, i) for i in range(epochs)]))
# 2.2: Initialize a pre-allocated DataFrame and assign the MultiIndex according to the meta data from each model.
df = pd.DataFrame(np.zeros((sum(numRows), 4), dtype=float), columns = ["loss", "accuracy", "val_loss", "val_accuracy"])
new_index = pd.MultiIndex.from_tuples(newIndex, names = ('Model_Type', 'job_ID', 'neurons', 'learning_rate', 'batch_size','epoch'))
df.index = new_index
print(df)
# 2.3: Now, populate the data frame by assigning the values from the CSV into their
# respective location in the pre-allocated DataFrame.
# AND
# 2.4: Query the model's graphic results (loss v. epochs and accuracy v. epochs graphs).
from matplotlib import pyplot as plt
from matplotlib import image as mpimg
prevNum = 0
for i in range(len(dirList)):
numMax = numRows[i]+prevNum
dirI = dirList[i]
df_temp = pd.read_csv(dirI+"/model_history.csv")
df.iloc[prevNum:int(numMax)] = df_temp
# Comment the next few lines out if intending to print the data frames here
# indexT = newIndex[prevNum:numMax]
# indexT = pd.MultiIndex.from_tuples(indexT, names = ('Model_Type', 'job_ID', 'neurons', 'learning_rate', 'batch_size','epoch'))
# df_temp.index = indexT
# print(df_temp)
# Now, show the graphs
image = mpimg.imread(dirI + "/loss_acc_plot.png")
currMetaData = newIndex[i]
currMetaData = newIndex[prevNum]
titleI = currMetaData[1]+ ": "+currMetaData[0] + " "+ currMetaData[2]+"N_lr"+str(currMetaData[3]) + "_bs"+str(currMetaData[4])+ "_e"+str(numRows[i])
plt.title(titleI)
fig = plt.imshow(image)
plt.axis('off')
fig.axes.get_xaxis().set_visible(False)
fig.axes.get_yaxis().set_visible(False)
plt.show()
plt.close()
prevNum = numMax
## Load in the model, though for this case, we do not need this
# for dirI in dirList:
# model_1H = keras.saving.load_model(dirI + "/model_weights.h5")
Step 3: Validation phase: Visualization of training data
1) Inspect if the trainings did not go as expected
2) Visually inspect for anomalies
3) Visually or numerically check for convergence (e.g. check the last 4-5 epochs, what the slope is like; any fluctuations?)
This is the step where one can easily look at the graphics and determine hyperparameters that produce bad results. For example, the graphic produced from the model with 0.1 learning rate does not appear to produce good results. Both the training loss and validation loss do not exhibit the typical trend. It does not appear to stabilize at all during the 30 epochs.
Step 4: Analysis phase: Visualizing the results
Let’s look at the last epoch data for each model. Since the models only varies one hyperparameter at a time, let’s create a graphical representation of the last epoch metrics vs. varying hyperparameter.
# Create last epoch DataFrame.
# Filter the DataFrame to only get the rows where the index for epoch is equal to the maximum number of epochs-1.
last_epoch_data = df.loc[(df.index.get_level_values('epoch')==max(numRows)-1)]
print(last_epoch_data)
# Make the DataFrame more readable
last_epoch_data2 = last_epoch_data.reset_index(drop=False)
loss accuracy val_loss val_accuracy
neurons
80 0.007796 0.998654 0.011362 0.998425
40 0.016035 0.996434 0.017410 0.996375
18 0.045203 0.990570 0.043723 0.990589
12 0.070112 0.985663 0.068991 0.985389
8 0.183243 0.952651 0.183629 0.950912
4 0.463814 0.907489 0.460035 0.910356
1 1.945121 0.319691 1.947505 0.320510
2 1.051278 0.682108 1.061323 0.682804
INSERT GRAPHICS
INSERT DISCUSSIONS FROM PREV EPISODE?
Key Points
How scripting works by converting the notebook to job scripts
Build a simple toolset/skillset to create, launch, and manage the multiple batch jobs.
Use this toolset to obtain the big picture result after analyzing the entire calculation results as a set.
Use Jupyter notebook as the workflow driver instead of using it to do the heavy-lifting computations on it.